By Syed Moiz Ahmed
Contribution participating in the INPST 2018 Science Communication Awards contest.
With the rapid increase in our understanding of disease biology, it is quite evident that treatment outcomes are dependent on patient molecular profiles. The focus is now to get the right treatment to the right patient allowing for greater safety and efficacy. The authors have shed light on a molecular biomarker that could potentially be used in this new treatment paradigm.
Chemotherapy has been used for treating cancer patients for more than 40 years. It is however associated with several side effects, and tumor recurrence is still a major unsolved issue. To prevent relapse and improve patient survival, current treatment regimens include chemotherapy alone or in combination with surgery/radiotherapy/more recently developed immunotherapy.
Different cancer types require different treatment options with one goal, that is to thwart tumor progression and prevent tumor recurrence or activate the immune response of the patient. Promising new therapies involving new drug targets are also being fast-tracked in ongoing clinical trials in the hope to improve patient survival.
One such modality of treatment involves the use of chemotherapeutic drugs that target topoisomerases. Cells are equipped with two major types of topoisomerases, namely topoisomerase I (TOP1) and topoisomerase II (TOP2). Current inhibitors to these enzymes (FDA-approved) are mostly plant alkaloids with several modifications to reduce side effects. TOP1 inhibitors called camptothecins (CPT), derived from the bark of the Chinese tree Camptotheca acuminate were among the first and most widely used drugs due to its potency to hinder the growth of leukemia cells. In particular, Irinotecan and Topotecan, two CPT derivatives that are more stable compounds, are extensively used in the treatment of colorectal and ovarian cancers respectively (Pommier, 2009). TOP2 inhibitors such as Etoposide and Teniposide are semisynthetic derivatives from the rhizome of Podophyllum peltatum and are used in treating a wide range of cancers today.
Topoisomerases and their Relevance to Genome Stability
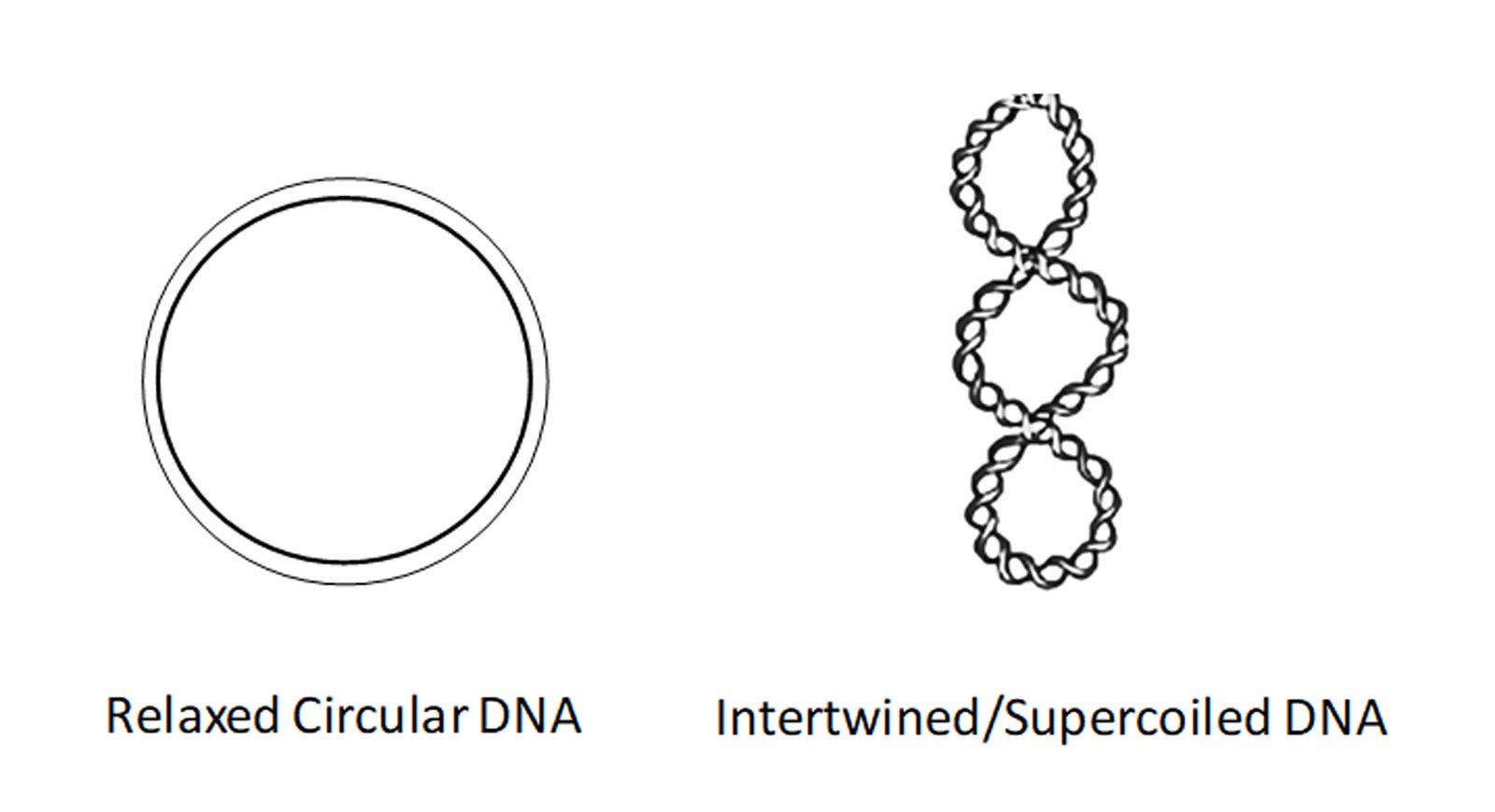
Topological states of DNA (imagine DNA as a rope which is intertwined, and its untangling is impossible due to lack of a free end) control almost all known processes key to a cell’s survival.
In cancer cells, this situation is more dangerous as the intertwining dramatically increases due to their uncontrolled cell division (Slaymaker et al., 2013). Topoisomerases are crucial enzymes that are equipped to modulate these DNA topological states, thereby releasing torsional tension (imagine the rope is cut, providing a free end that allows us to rotate and untangle it). Intertwined DNA can terminate replication/transcription in its tracks leading to single/double strand breaks and cell death. Although a highly underappreciated and technically very challenging process to detect and study, DNA topology as a scientific field has garnered interest as a prominent force that drives life as we know it.
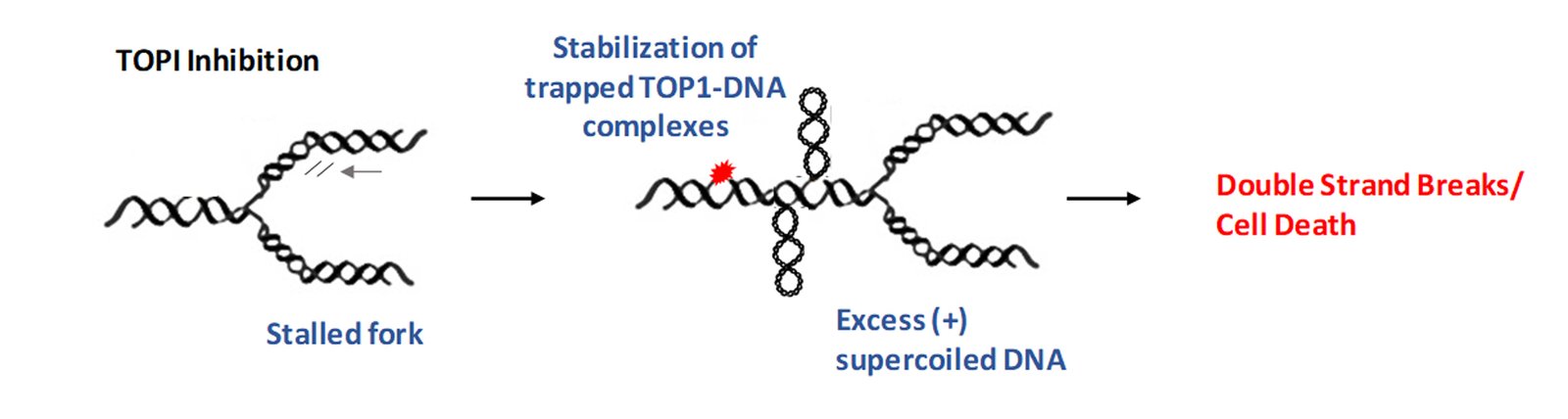
Topoisomerases remove such complex DNA structures and allow smooth progression of DNA replication/transcription. However, these enzymes generate a break in the DNA strand which allows for free rotation of the cut end leading to untangling of intertwined DNA. Topoisomerase inhibitors exploit this process of generating a break on the DNA. In particular, TOP1 inhibitors, such as camptothecin (CPT) derivatives stabilize a key intermediate step and trap the topoisomerase enzyme in a complex with DNA, preventing DNA relaxation. This potentiates two scenarios not exclusive to each other. First, the trapped enzyme complex hinders replication fork movement leading to fork stalling, and if not resolved quickly this can lead to the generation of double-strand breaks (DSBs) (Tomicic and Kaina, 2013). Second, the inhibition of TOP1 activity can lead to excess intertwining of DNA (also called supercoiled DNA), which then potentiates the replication fork to collapse, leading to genome instability as shown below (Menin et al., 2018).
HMGA2 – Master Regulator of Replication Stress
Any cellular system in nature is inclined to adopt more direct and energy efficient processes to overcome potential challenges. It is widely believed that cancer cells are also equipped to tackle genome-wide challenges thrown at them particularly during chemotherapy. With DNA damage response and repair systems predominantly slow, factors working at the ground level should exist. Prof. Peter Dröge and his team discovered that the chromosomal protein factor HMGA2 (high mobility group AT Hook 2) prevents replication fork collapse in cancer cells and human embryonic stem cells. Their paper published in Cell Reports showed that the protein utilizes its three DNA binding domains to bind at stalled forks and prevent DNA strand breaks in human, bacterial and yeast cell systems (Yu et al., 2014). Additionally, they also found that HMGA2 binds with high affinity to supercoiled DNA, which was reported recently in Scientific Reports, providing insights into how HMGA2 could regulate topological problems in cancer cells (Zhao et al., 2017).
However, some key questions remain. Although topoisomerases are useful chemotherapeutic targets, cancer cells can surprisingly manage these topological challenges leading to futile chemotherapeutic responses in patients.
HMGA2 regulates topological problems in cancer cells
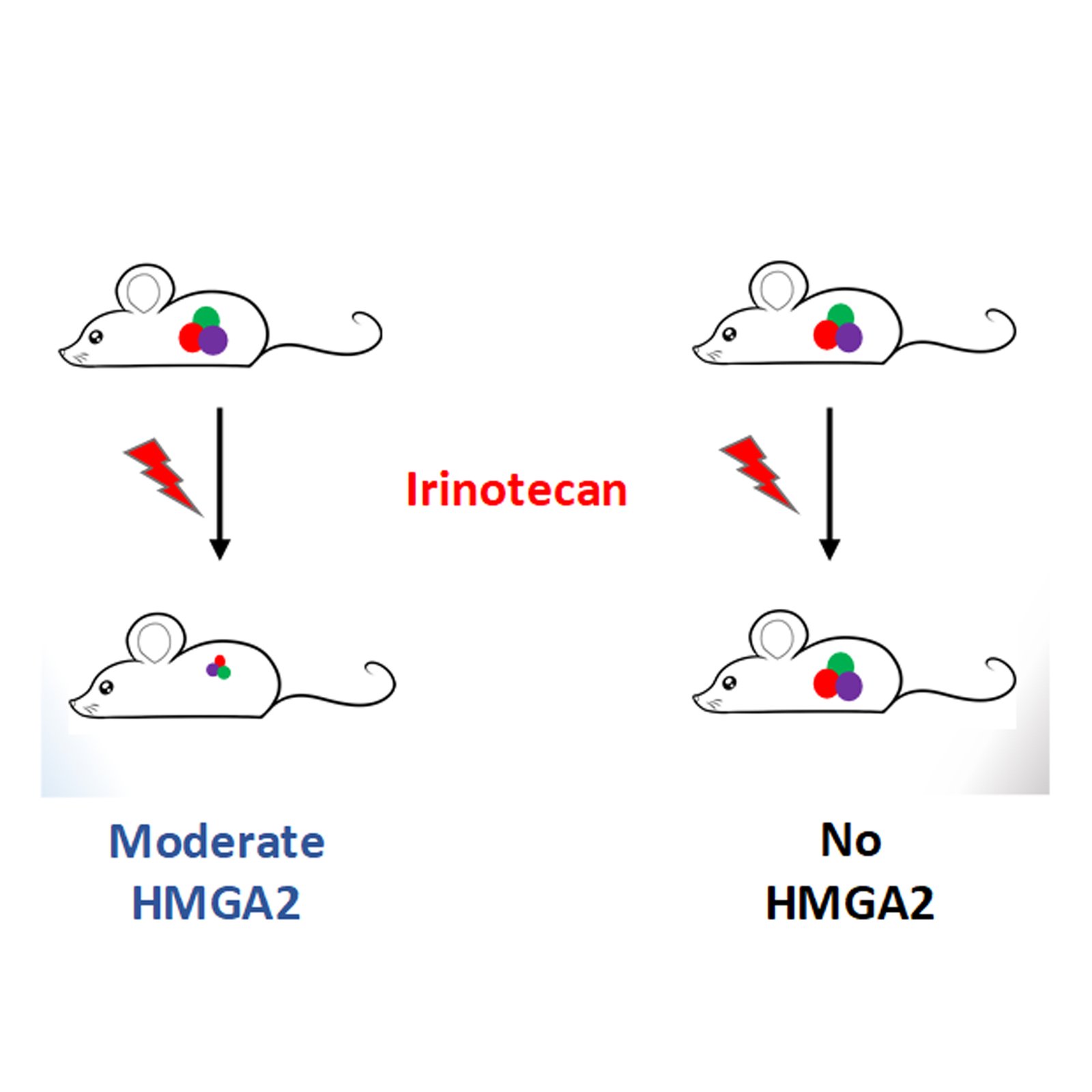
To answer this question, Prof. Peter Dröge and his team utilized the camptothecin derivative Irinotecan to inhibit topoisomerase I in HMGA2-expressing cancer cells. In particular, they used a more potent, active metabolite of irinotecan called SN38. As it turns out quite surprisingly, HMGA2 expression levels in cancer cells determine sensitivity to SN38. They found that cancer cells expressing higher levels of the protein are resistant to the drug, whereas cells that express comparatively moderate levels are sensitized to SN38. They argue that such an effect is due to HMGA2 binding/constraining excess supercoiled DNA generated as a result of TOP1 inhibition and that HMGA2 interferes with drug binding to TOP1 leading to such a dynamic response. At high HMGA2 levels, the protein most likely competes with TOP1 binding to DNA, and hence low levels of TOP1-DNA complexes are generated. At moderate levels, the protein increases SN38-TOP1-DNA interactions leading to higher levels of complex formation and DNA damage.
To strengthen their study, the team further analyzed human tumor samples in a classic experiment called ‘xenografting’, where small pieces of tumors are engrafted into immunocompromised mice and allowed to grow for up to a month. Irinotecan was used to treat these mice and tumor growth was monitored. Such a study mimics tumor growth in humans and allows us to study tumor response to chemotherapeutic drugs. Consistent with the cell line data, they found that tumors that express moderate levels of HMGA2 reduced in size with Irinotecan treatment, pointing to increased DNA damage which ultimately leads to cell death. Interestingly, tumors that did not express HMGA2 continued to grow after Irinotecan treatment. Such a finding holds excellent promise as HMGA2 expression levels could guide clinicians in the use of Irinotecan as first-line therapy.
The authors suggest that tumors that express moderate amounts of HMGA2 can be selected for Irinotecan treatment as most tumors regress whereas tumors that express comparatively higher amounts of HMGA2 could be chosen for other effective strategies, as shown above, allowing a sort of personalized medicine for patients enduring a painful journey in their cure for cancer.
Conclusion
Overall, HMGA2 could be utilized as a predictive molecular marker in treating cancer patients, especially those that are under camptothecin based chemotherapy. As an ongoing effort to improve chemotherapeutic response, new camptothecin-like inhibitors are being currently developed and investigated.
References
MENIN, L., URSICH, S., TROVESI, C., ZELLWEGER, R., LOPES, M., LONGHESE, M. P. & CLERICI, M. 2018. Tel1/ATM prevents degradation of replication forks that reverse after topoisomerase poisoning. EMBO Rep, 19.
POMMIER, Y. 2009. DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem Rev, 109, 2894-902.
SLAYMAKER, I. M., FU, Y., TOSO, D. B., RANATUNGA, N., BREWSTER, A., FORSBURG, S. L., ZHOU, Z. H. & CHEN, X. S. 2013. Mini-chromosome maintenance complexes form a filament to remodel DNA structure and topology. Nucleic Acids Res, 41, 3446-56.
TOMICIC, M. T. & KAINA, B. 2013. Topoisomerase degradation, DSB repair, p53 and IAPs in cancer cell resistance to camptothecin-like topoisomerase I inhibitors. Biochim Biophys Acta, 1835, 11-27.
YU, H., LIM, H. H., TJOKRO, N. O., SATHIYANATHAN, P., NATARAJAN, S., CHEW, T. W., KLONISCH, T., GOODMAN, S. D., SURANA, U. & DROGE, P. 2014. Chaperoning HMGA2 protein protects stalled replication forks in stem and cancer cells. Cell Rep, 6, 684-97.
ZHAO, X., PETER, S., DROGE, P. & YAN, J. 2017. Oncofetal HMGA2 effectively curbs unconstrained (+) and (-) DNA supercoiling. Sci Rep, 7, 8440.
Keywords: predictive molecular marker in treating cancer, DNA damage, DNA intertwining, replication fork, high mobility group AT Hook 2, HMGA2, supercoiled DNA, Teniposide, DNA replication/transcription, cell death, predictive molecular markers, cancer cells, Etoposide, single/double strand breaks, Topotecan, Irinotecan, camptothecins, topoisomerases, cancer, chemotherapy.
Join for free INPST as a member
The International Natural Product Sciences Taskforce (INPST) maintains up-to-date lists with conferences, grants and funding opportunities, jobs and open positions, and journal special issues with relevance for the area of phytochemistry and food chemistry, pharmacology, pharmacognosy research, and natural product science.
Leave a comment:
[contact-form][contact-field label=”Name” type=”name” required=”true” /][contact-field label=”Email” type=”email” required=”true” /][contact-field label=”Website” type=”url” /][contact-field label=”Message” type=”textarea” /][/contact-form]