Abstract:
Unknown metabolites represent a bottleneck in untargeted metabolomics research. Ion mobility-mass spectrometry (IM-MS) facilitates lipid identification because it yields collision cross section (CCS) information that is independent from mass or lipophilicity. To date, only a few CCS values are publicly available for complex lipids such as phosphatidylcholines, sphingomyelins, or triacylglycerides. This scarcity of data limits the use of CCS values as an identification parameter that is orthogonal to mass, MS/MS, or retention time. A combination of lipid descriptors was used to train five different machine learning algorithms for automatic lipid annotations, combining accurate mass (m/z), retention time (RT), CCS values, carbon number, and unsaturation level. Using a training data set of 429 true positive lipid annotations from four lipid classes, 92.7% correct annotations overall were achieved using internal cross-validation. The trained prediction model was applied to an unknown milk lipidomics data set and allowed for class 3 level annotations of all features detected in this application set according to Metabolomics Standards Initiative (MSI) reporting guidelines.
Reference:
Blaženović I, Shen T, Mehta SS, Kind T, Ji J, Piparo M, Cacciola F, Mondello L, Fiehn O. Increasing Compound Identification Rates in Untargeted Lipidomics Research with Liquid Chromatography Drift Time-Ion Mobility Mass Spectrometry. Anal Chem. 2018 Sep 18;90(18):10758-10764. doi: 10.1021/acs.analchem.8b01527.
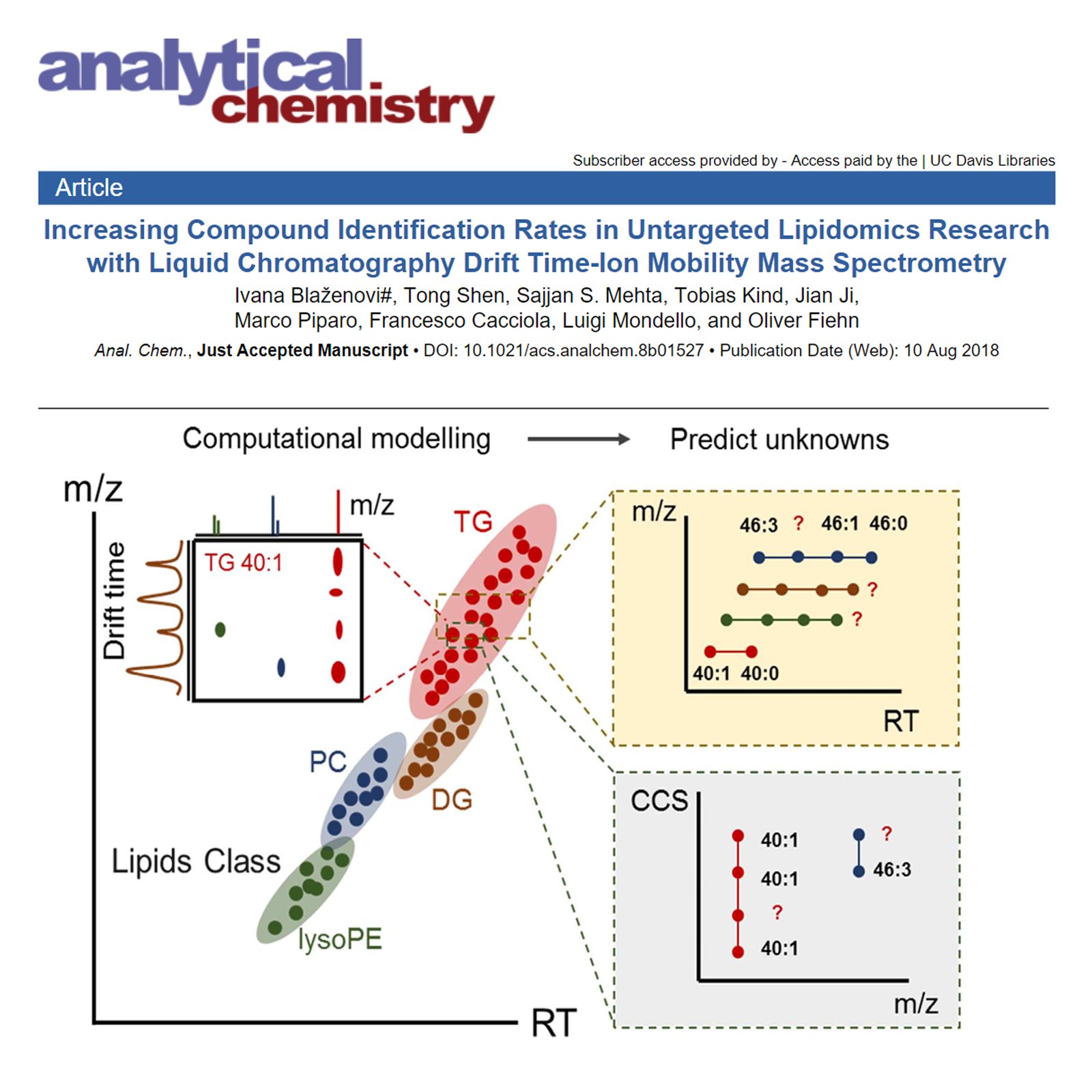
Keywords: increasing compound identification rates, untargeted lipidomics research, liquid chromatography drift time-ion mobility mass spectrometry, milk lipidomics, collision cross section (CCS), untargeted metabolomics.
Join for free INPST as a member
The International Natural Product Sciences Taskforce (INPST) maintains up-to-date lists with conferences, grants and funding opportunities, jobs and open positions, and journal special issues with relevance for the area of phytochemistry and food chemistry, pharmacology, pharmacognosy research, and natural product science.